A high-resolution correlative light and X-ray 3D cryo-imaging pipeline for cells and tissue at near-physiological conditions
- Abstract number
- 393
- Presentation Form
- Contributed Talk
- DOI
- 10.22443/rms.mmc2023.393
- Corresponding Email
- [email protected]
- Session
- Multiscale and Correlative Microscopy Approaches to Microanalysis and Spectroscopy
- Authors
- Dr Archana Jadhav (1), Dr Chidinma Okolo (1), Dr Kamal Nahas (1), Mr Thomas Fish (1), Mr Adam Taylor (1), Dr Maria Harkiolaki (1)
- Affiliations
-
1. Diamond Light Source Ltd
- Keywords
soft X-ray tomography, structured illumination microscopy, X-rays, correlative imaging
- Abstract text
High resolution imaging of the cellular universe in recent years has underpinned a revolution in high-content volume data accumulation resulting in a new understanding of cellular organisation and behaviour. Among the highlights of microscopy developments in cellular imaging has been the emergence of new correlative imaging modalities that incorporate Soft X-ray Tomography (SXT) of cells and tissue that have been cryogenically prepared in a way that retains native structures and architecture without sample damage or artefact creation. At the correlative cryo-imaging beamline B24 of the UK synchrotron we have devised and refined such a correlative imaging platform to a high level of automation and throughput [1] by focusing not only on technique development but perhaps more importantly on accessibility and ease or use.
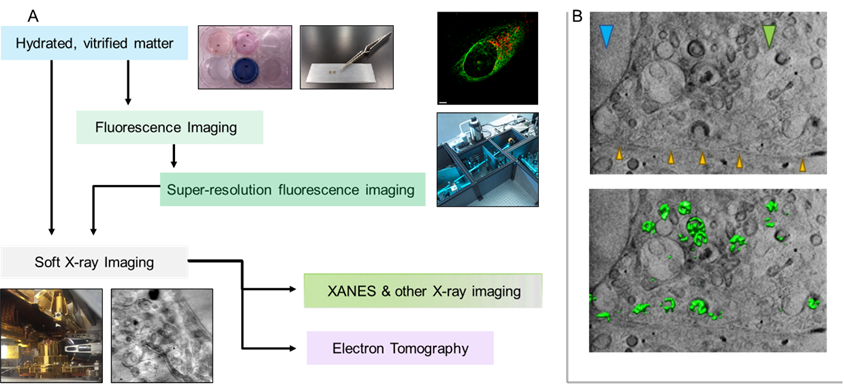
SXT is a high-resolution 3D mesoscale imaging technique for cells and tissues up to 12 μm in thickness in fully hydrated state without employing chemical fixation, sectioning, or milling, or the use of contrast-enhancing agents. Under cryogenic conditions vitrified samples are irradiated with “water-window” X-rays to achieve lateral resolution of structures within cells to 25 nm depending on the optical setup used in SXT microscope. Ongoing development in correlative imaging has led to new ways of combining SXT technique with other corresponding methods such as spectroscopy, X-ray fluorescence microscopy, and electron microscopy for correlative imaging. At B24, a 3D super resolution fluorescence microscope based on structured illumination microscopy (SIM) principles was developed as a complementary tool to match molecular localisation with high-definition structural data acquired in SXT under cryogenic conditions leading to the development of a unique same-sample correlative light and X-ray tomography (CLXT) platform for biological research. SIM allows the acquisition of high-resolution fluorescence data with multi-channel image registration using commonly used fluorophores in thick vitrified samples.
A fully commissioned CLXT workflow has been developed at B24 beamline (Figure 1) [1]. The CLXT experimental workflow is adaptable for a broad range of biological specimens such as multi-species, adherent or suspension cell cultures (primary cells and immortalized cell lines), bacteria, parasites, algae, fungi, and viruses. Transmission electron microscopy gold grids are used as sample carriers and biological samples are either cultured or deposited on them. Live cells can then be labelled using different fluorescent cell trackers. Vitrification is at the heart of CLXT workflow, where biological samples on the TEM grids are immobilized by cryofixation using the rapid plunge freezing method. Gold nanoparticles are added as image registration markers and excess of liquid is removed by the blotting on filter paper. Cryofixed samples are then assessed for vitrification quality using wide-field fluorescence imaging and mapped in bright-field and fluorescence before loading into the 3D cryo-SIM. The same sample is then loaded into the soft X-ray microscope and SXT data is collected from the same cell and the same regions of interests (ROIs) where 3D super resolution fluorescence data has been collected previously. SXT raw data are collected as series of 2D projection images of X-rays through the sample as it is rotated through a range of angles and finally reconstructed into 3D tomograms. The final step of CLXT workflow is data analyses of information generated through cryo-SIM and cryo-SXT, which can then be used for correlating functional, chemical colocalization with structural information using an open-source software plugin (ec-CLEM) in ICY [2] and for quantitative outputs on features of promising interests through segmentation analysis [3].
Here we will highlight representative B24 data from current biomedical areas and disease developments in both methodology and applications alongside new results in infection and immunology areas that have been only recently discovered through the use of the B24 platform. Access routes, best practices, and data analysis tool kits will also be presented to allow an in-depth understanding of both the applicability and the potential of correlative 3D cryo-imaging with soft X-rays at a synchrotron setting.
The cryogenic CLXT platform of B24 has been proven to be a powerful tool by employing cutting-edge microscopic techniques of SIM and SXT to study viruses, bacteria, parasites, fungi, and different types of cells (can be treated with different chemicals) in their near-physiological state and enabled research community to derive meaningful functional and structural information explicitly. This platform is easily accessible to the global scientific research community and both microscopes offer rapid acquisitions of samples to provide high-resolution information making it useful for high throughput high-content volume imaging.
The SXT platform can also be paired with other techniques such as electron tomography, FIB-SEM, spectroscopy, X-ray fluorescence microscopy, and others to obtain multi-fold information from the specimen continue to provide innovative ways towards increasing the dimensionality of data obtained through correlative imaging to interlink different disciplines of science.
Figure 1. Super resolution fluorescence and soft X-ray imaging acquired from vitrified cell using the same-sample correlative cryo-imaging platform of B24 (A). X-ray tomographic z-slice obtained from the infected U2OS cell elucidates colocalization of virus (green fluoresce) in different cytoplasmic vesicles revealed in correlation analysis (B). Cellular features are identified by arrows: orange for cell membrane, blue for nucleus, and green for mitochondria.
- References
1. Okolo CA, Kounatidis I, Groen J, Nahas KL, Balint S, Fish TM, Koronfel MA, Cortajarena AL, Dobbie IM, Pereiro E, Harkiolaki M (2021) Sample preparation strategies for efficient correlation of 3D SIM and soft X-ray tomography data at cryogenic temperatures. Nat Protoc. 16:2851-2885. https://doi.org/10.1038/s41596-021-00522-4
2. Vyas N, Kunne S, Fish TM, Dobbie IM, Harkiolaki M, Paul-Gilloteaux P (2021). Protocol for image registration of correlative soft X-ray tomography and super-resolution structured illumination microscopy images. STAR Protoc. 2:100529. https://doi.org/10.1016/j.xpro.2021.100529
3. Luengo I, Darrow MC, Spink MC, Sun Y, Dai W, He CY, Chiu W, Pridmore T, Ashton AW, Duke EMH, Basham M, French AP (2017) SuRVoS: Super-Region Volume Segmentation workbench. Journal of Structural Biology. 198:43-53. https://doi.org/10.1016/j.jsb.2017.02.007