Using advanced microscopy and microfluidics to study evolutionary processes involved in the emergence of AMR
- Abstract number
- 72
- Presentation Form
- Poster & Flash Talk
- DOI
- 10.22443/rms.mmc2023.72
- Corresponding Email
- [email protected]
- Session
- Poster Session Two
- Authors
- Dr Raveen Tank (1), Dr Rok Krasovec (1)
- Affiliations
-
1. The University of Manchester
- Keywords
PALM
AMR
TIRF
Microfluidics
E.coli
DNA Repair
Mutation rate
- Abstract text
Bacteria have an extraordinary capacity to grow, reproduce and evolve. Most bacteria are haploid for the vast majority of their genes, so the probability at which mutations occur in these genes critically affect the rate of bacterial growth and adaptation. Studying mutations is thus of fundamental importance for the understanding of bacterial evolution and emergence of a key global challenge of antimicrobial resistance. Previous work has demonstrated that spontaneous mutations rates can vary plastically and can be dependent on environmental factors 1. It is know that an organisms’ mutation rates are associated with the density to which they grow, a higher population density is associated with lower mutation rates 2, density associated mutation rate plasticity (DAMP).
Super resolution microscopy has been used to study bacteria in order to understand mechanisms involved in antimicrobial resistance3. Here we continue to use advanced microscopy techniques to understand mechanisms involved in density-associated mutation rate plasticity. We investigate how a DNA repair protein, MutS, interacts with the DNA in accordance with the cell density. We use a custom built microfluidic device to confine Escherichia coli cells within chambers varying in geometry, allowing cells to grow into different density. The device is imaged using photo activated light microscopy (PALM) and total internal reflection fluorescence (TIRF) microscope, which enables visualisation of single MutS molecules. As well as looking at different density conditions, we also have genetic knockouts where a number of DNA repair proteins are knocked out, thus allowing us to identify the mechanisms responsible for DAMP.
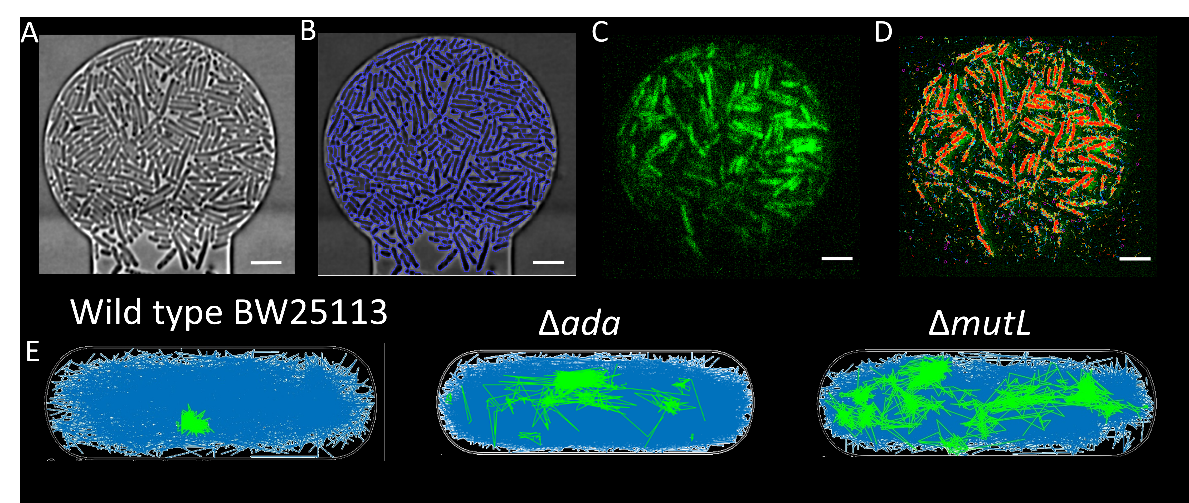
Figure 1A shows the pipeline of data processing, where a phase contrast image of cells in chambers is captured, the image is then segmented to show the cell outline. Data showing the fluorescence video of MutS DNA repair proteins movement is then processed to show track trajectory. Using the cell segmentation and the track information, the track displacement of MutS can then be compared in different density conditions and also in the different genetic knockouts.
Figure 1A) Cells packed into a chamber at high density, B) cell segmentation giving clear boundaries to the cells, C) fluorescence image showing signal from MutS DNA repair protein, D) red tracks showing movement of the DNA repair proteins. E) tracks showing the difference in track trajectory in DNA repair protein knock out strains, where mutL and ada have more stationary tracks (shown in green).
1. Krašovec R, Belavkin R v., Aston JAD, et al. Mutation rate plasticity in rifampicin resistance depends on Escherichia coli cell-cell interactions. Nat Commun. 2014;5. doi:10.1038/ncomms4742
2. Krašovec R, Richards H, Gifford DR, et al. Spontaneous mutation rate is a plastic trait associated with population density across domains of life. PLoS Biol. 2017;15(8):1-19. doi:10.1371/journal.pbio.2002731
3. Banaz N, Mäkelä J, Uphoff S. Choosing the right label for single-molecule tracking in live bacteria: Side-by-side comparison of photoactivatable fluorescent protein and Halo tag dyes. J Phys D Appl Phys. 2019;52(6). doi:10.1088/1361-6463/aaf255
- References
1. Krašovec R, Belavkin R v., Aston JAD, et al. Mutation rate plasticity in rifampicin resistance depends on Escherichia coli cell-cell interactions. Nat Commun. 2014;5. doi:10.1038/ncomms4742
2. Krašovec R, Richards H, Gifford DR, et al. Spontaneous mutation rate is a plastic trait associated with population density across domains of life. PLoS Biol. 2017;15(8):1-19. doi:10.1371/journal.pbio.2002731
3. Banaz N, Mäkelä J, Uphoff S. Choosing the right label for single-molecule tracking in live bacteria: Side-by-side comparison of photoactivatable fluorescent protein and Halo tag dyes. J Phys D Appl Phys. 2019;52(6). doi:10.1088/1361-6463/aaf255