Investigating the influence of cell shape on intra-colony channel architecture by fractal analysis of confocal microscopy biofilm images
- Abstract number
- 389
- Presentation Form
- Contributed Talk
- Corresponding Email
- [email protected]
- Session
- Microbial Imaging
- Authors
- Ms Beatrice Bottura (1), Prof. Paul Hoskisson (1), Prof. Gail McConnell (1)
- Affiliations
-
1. University of Strathclyde
- Keywords
Confocal microscopy, biofilms, fractal geometry
- Abstract text
We present a morphological analysis of intra-colony channels in E. coli biofilms imaged by confocal laser scanning microscopy. We quantified channel morphology using the fractal geometry quantities of box-counting dimension and lacunarity, which we compared across six E. coli strains with differing cell shapes.
Nutrient-transporting channels are found throughout the volume of mature E. coli biofilms. They have the ability to transport small fluorescent molecules and they can reaggregate after mechanical disruption (Rooney et al. 2020). It has recently been found that the morphology and distribution of these channels is impacted by environmental growth conditions, such as nutrient availability and substrate stiffness (Bottura et al. 2022).
While the effect of cell morphology on internal biofilm structure has been studied in simulation and experiments (Smith et al. 2017), its specific role on intra-colony channel architecture is unknown. We hypothesised that cell shape affects the 3D distribution of these channel networks inside mature biofilms, and in particular their fractal geometry.
We tested this hypothesis by imaging the structure of biofilms formed by six E. coli strains with different cell morphologies. Five single gene-knockout mutants from the Keio collection (Baba et al. 2006) were chosen for their altered cell phenotype: ΔamiA and ΔguaB (long cells), ΔompR (fat cells), ΔtolB (chains of cells) and ΔydgD (fat cells). The genotype of each mutant strain was checked by PCR and sequencing, and cell phenotypes were confirmed by phase contrast microscopy. The wild-type parental strain for the Keio collection, BW25113, was used as a reference for morphological measurements.
After transforming all six strains with the GFP fluorescent plasmid rpsM::gfp, we grew biofilms on agar substrates and imaged them with an Olympus FV1000 fluorescence microscope using a 10x/0.4 air objective lens. We used a combination of fluorescence microscopy and open-source image analysis software to quantify the variation in internal biofilm architecture via fractal geometry measurements. Using the ridge detection plugin (Wagner & Hiner, 2017) on ImageJ, we created a binary mask of the channels inside each biofilm image. We used the twombli plugin (Wershof et al. 2021) to calculate the fractal geometry quantities of lacunarity and box-counting fractal dimension from each binary mask. Ten biofilm images were analysed for each strain.
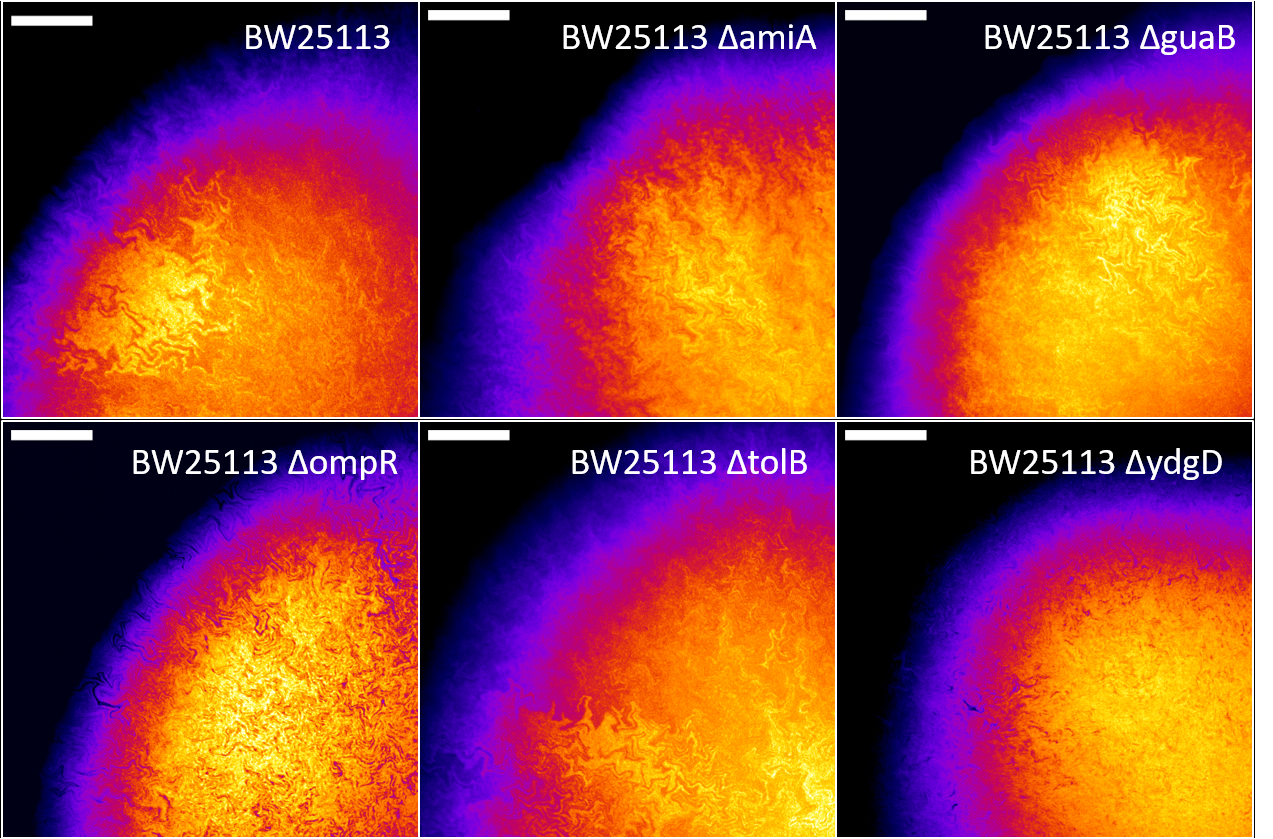
Confocal fluorescence microscopy images of the biofilms formed by the six E. coli strains revealed a complex network of fractal intra-colony channels (Figure 1).
Figure 1: Biofilms formed by the Keio parental strain BW25113 and five cell shape mutants, showing complex fractal networks of intra-colony channels. Each image is a maximum intensity projection of a z-stack with slice spacing of 3 μm, acquired with an Olympus FV1000 confocal microscope. Scale bars: 250 μm.
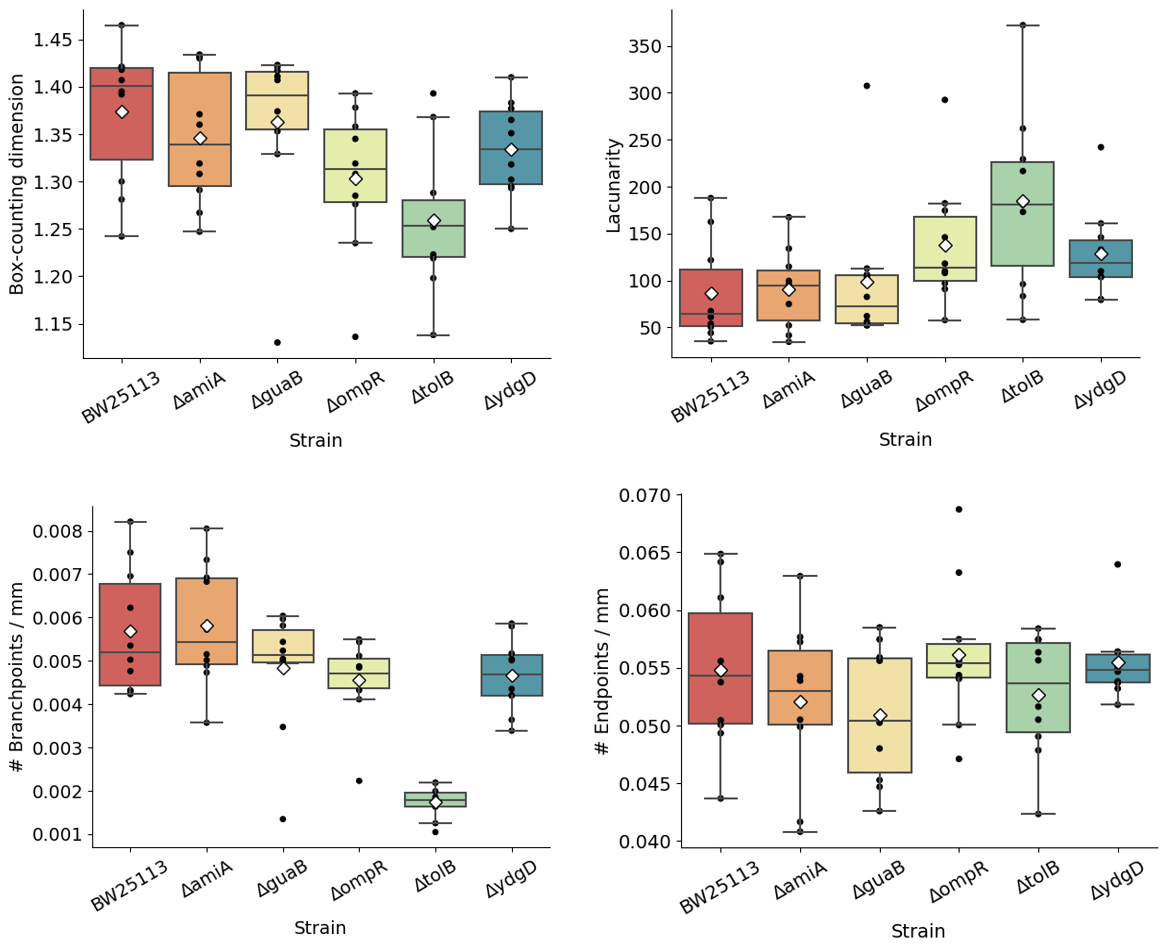
The fractal geometry of these channels was analysed on FIJI by measuring box-counting fractal dimension and lacunarity. We observed that the two quantities are inversely related, and that intra-colony channel architecture inside the biofilms is closely linked to cell phenotype (Figure 2). Strains with the same phenotype, e.g. ΔamiA and ΔguaB (long cells) and ΔompR and ΔydgD (fat cells), have comparable fractal measurements. Fractal measurements also differ between strains with different phenotypes. For example, channels from the chain-forming ΔtolB have a lower box-counting dimension (1.259) and form biofilms with higher lacunarity (185.3), whereas the fat cells from ΔompR and ΔydgD have higher box-counting dimension (1.303 and 1.334 respectively) and lower lacunarity (137.7 and 128.7 respectively). Biofilms formed by long cells, ΔamiA and ΔguaB, had a box-counting dimension of 1.346 and 1.362 respectively, and lacunarity of 90.9 and 98.9 respectively - these values are similar to those of the parental strain BW25113, which has box-counting dimension 1.374 and lacunarity 86.7.
Fractal domains have been previously observed in bacterial cocultures of both E. coli and B. subtilis, where they result from mechanical instability during colony expansion (Rudge et al. 2013, Amor et al. 2017). Simulations of diffusion-limited aggregation in B. subtilis and S. cerevisiae also show fractal-like growth under nutrient limitation (Ginovart et al. 2002). Our findings show that cell shape is directly linked to intra-colony channel architecture in E. coli biofilms.
Figure 2: Box plots showing box-counting fractal dimension (left) and lacunarity (right) for Keio mutant biofilms. Average values are shown as white diamonds, whereas boxes represent the interquartile range (with median values shown as horizontal lines inside each box). Whiskers enclose the 5th and 95th data percentiles.
In conclusion, we have analysed the internal morphology of E. coli biofilms formed by cells with normal, long, chained and fat phenotypes using confocal microscopy and an open-source image analysis pipeline. By measuring the fractal geometry parameters of box-counting dimension and lacunarity, we showed that the fractal organisation of channels inside the biofilm are dictated by the cellular phenotype. The image analysis pipeline used in this work can be easily adapted to the study of filamentous internal biofilm structures, provided a high enough contrast between them and the rest of the biofilm.
- References