Using Deep Learning based reconstruction techniques to improve image quality and reduce scan times of plant biology samples scanned with X-ray microscopes
- Abstract number
- 74
- Presentation Form
- Poster & Flash Talk
- DOI
- 10.22443/rms.mmc2023.74
- Corresponding Email
- [email protected]
- Session
- Artificial Intelligence
- Authors
- Dr Keith Duncan (1), Dr Rachna Parwani (2), Dr Rosy Manser (2)
- Affiliations
-
1. Donald Danforth Plant Science Center
2. ZEISS Research Microscopy Solutions
- Keywords
XRT, Computed Tomography, X-ray CT, XRM, X-ray microscopy, Deep learning reconstruction
- Abstract text
X-ray tomography (XRT) is a powerful technique for 3D visualisation of plant biology both above and below the soil line, whether using a synchrotron facility or a lab-based instrument. The non-destructive nature of XRT enables structural imaging without the need to physically disturb the plant, providing a unique approach to specimen assessment that is not practical or possible using light, laser, and electron microscopy platforms. Magnification in traditional lab-based XRT setup is limited by the source-sample-detector geometry of the instrument’s internal cabinet space. Conversely, the X-ray microscope (XRM) significantly increases available magnification by inserting scintillator-coated objective lenses into the beam path, allowing multiscale imaging that places high magnification scans within the context of the entire intact structure scanned at lower magnification1.
An important aspect of XRM imaging is computational reconstruction of the 3D volume from the hundreds or thousands of 2D digital radiographs collected during scanning. Most commercial XRM or XRT systems utilize the traditional Feldkamp-Davis-Kress (FDK) algorithm for cone-beam reconstruction. This can generate good quality images in a reliable reconstruction process, but the FDK method is sensitive to individual projection noise level, to photon starvation, and resulting images are prone to a variety of under-sampling artifacts. Consequently, a high number of projections, and/or long exposure times per projection are required for reducing image spurious noise and artifacts, which in turn means longer scans for high resolution data acquisition using high optical magnification objectives. For this reason, throughput of the 3D XRM process becomes a bottleneck, particularly when high resolution is needed across a large volume of the sample, for visualisation or representativity purposes.
A computational solution that substantially speeds up data acquisition whilst still providing high resolution 3D imaging would offer great benefit in terms of boosting productivity and gleaning new insights into plant specimens. Recent software advances have incorporated Deep Learning strategies into the reconstruction process that have increased XRM imaging throughput (>4 times) by reducing scan times without compromising image quality, and significantly increasing signal to noise ratio of the resulting data.
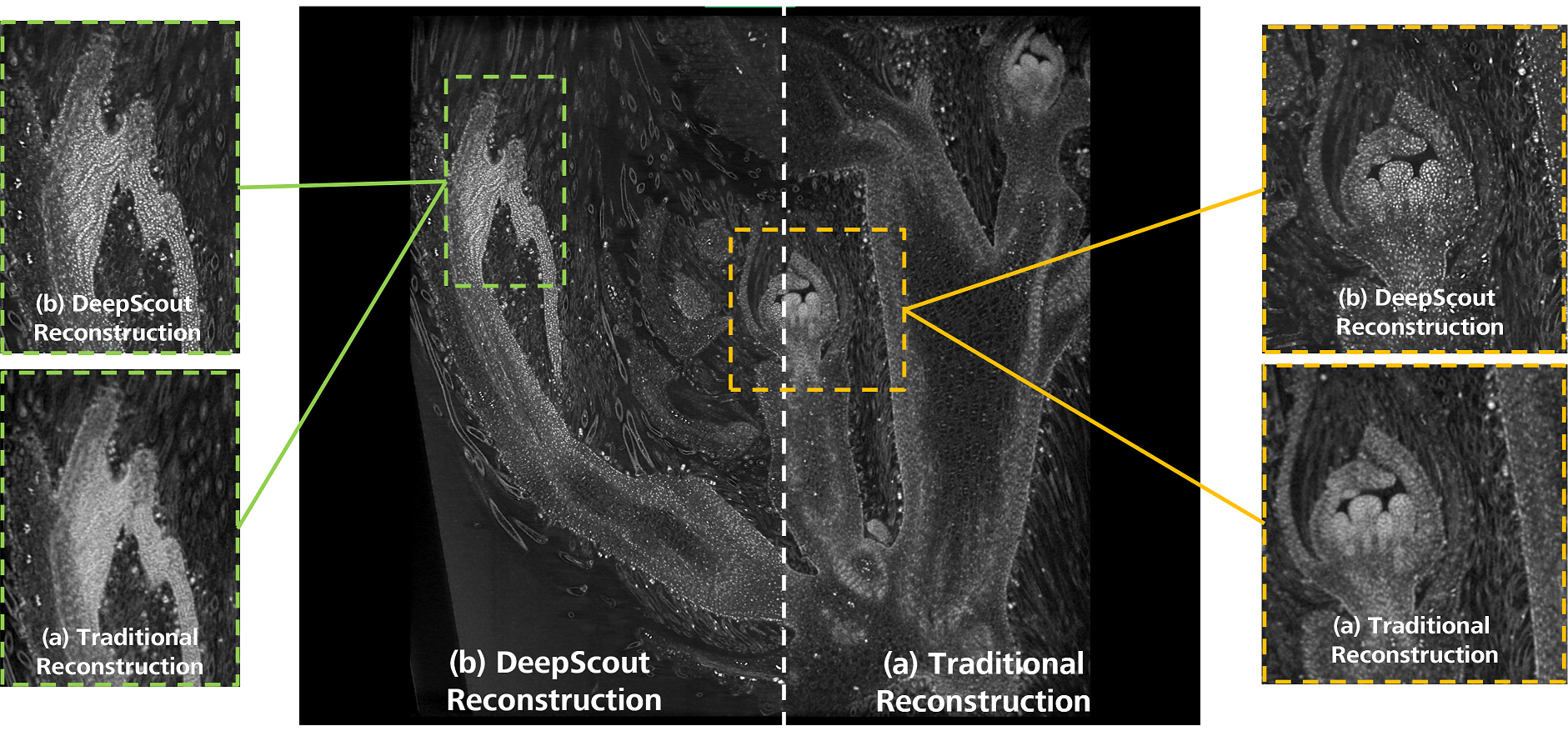
Here we demonstrate the use of a new Deep Learning method, DeepScout, capable of upscaling an image by a factor 5, by using a model developed from a convolutional neural network trained with a single high resolution internal tomography dataset registered with the lower resolution dataset (figure 1). Once trained, the model is applied to the larger, low resolution overview datasets to generate high resolution volumes of those regions. Several different plant tissues are imaged in this way to demonstrate the robustness of the approach for different tissues and densities. We demonstrate that this approach uncovers features in the bulk datasets that were previously unresolvable in the original data, providing exciting opportunities for exploring specimens in a way that was previously inaccessible.
Figure 1: Developing soybean flower imaged non-destructively using the 4X objective of the ZEISS Xradia Versa X-ray microscope. The same dataset was reconstructed twice using (a) traditional FDK reconstruction and (b) Deep Learning-based DeepScout reconstruction using a convolutional neural network model previously trained from a small region of high-resolution data captured with the 20X objective.
1. Duncan, K. E., Czymmek, K. J., Jiang, N., Thies, A. C., and Topp, C. N. (2022). X-ray microscopy enables multiscale high-resolution 3D imaging of plant cells, tissues, and organs. Plant Physiol. 188 (2), 831–845. https://doi.org/10.1093/plphys/kiab405
- References
Duncan, K. E., Czymmek, K. J., Jiang, N., Thies, A. C., and Topp, C. N. (2022). X-ray microscopy enables multiscale high-resolution 3D imaging of plant cells, tissues, and organs. Plant Physiol. 188 (2), 831–845. https://doi.org/10.1093/plphys/kiab405