Automation of imaging and control of the OpenFlexure Delta Stage using the OpenFlexure MATLAB client.
- Abstract number
- 219
- Presentation Form
- Poster
- DOI
- 10.22443/rms.mmc2021.219
- Corresponding Email
- [email protected]
- Session
- Poster Session 1
- Authors
- Dr Samuel McDermott (2), Dr Joel Collins (1), Mr Filip Ayazi (2), Dr Julian Stirling (1), Dr Richard Bowman (1), Prof Pietro Cicuta (2)
- Affiliations
-
1. University of Bath
2. University of Cambridge
- Keywords
microscope, 3D printed, open source, MATLAB, automation, dynamic differential microscopy
- Abstract text
The OpenFlexure Delta Stage microscope is a low-cost 3D-printed microscope, developed as a fully open-source project1. It is a variant of the OpenFlexure Microscope2. The OpenFlexure Delta Stage is a fully motorised microscope, uses a Raspberry Pi Camera for imaging, and is controlled using a Raspberry Pi. Its small size and low cost make it suitable for use on laboratory benches or in microbiological safety cabinets (MSCs). A MATLAB client has been developed which allows the OpenFlexure Delta Stage to be controlled from within a MATLAB session. These MATLAB classes can control the position of the stage, change the illumination and return images and videos in MATLAB native formats. This allows researchers to run automated experiments, including over several OpenFlexure Delta Stages simultaneously.
Figure 1: The OpenFlexure Delta Stage is a bench top microscope suitable for automated experiments.
The main body of the OpenFlexure Delta Stage (Figure 1) is a single, 3D-printed plastic flexure mechanism which can move the sample in three dimensions. It is actuated using three stepper motors. The optics module uses a Raspberry Pi camera v2 as the detector in an inverted microscope configuration. The illumination for transmitted light can be a simple white LED all the way to an addressable RGB LED grid. At the core of the Delta Stage is a Raspberry Pi, which controls these components. Loaded on the Raspberry Pi, the OpenFlexure Software is coded in Python and the Flask web application framework3. This controls the lower-level functionality of the microscope (for example sending commands to the motors and capturing images using the camera) and starts a web server, serving web APIs based on the W3C Web of Things interaction model4. This model defines properties, actions and events on the microscope which are mapped to HTTP URLs. The core functionality of the microscope is built into this API, and additional functionality (such as video recording) can be added with extensions, with their own API URLs5.
Because the OpenFlexure Software uses RESTful APIs, which are widely used across internet services6, it is possible to control the Delta Stage using HTTP requests from a variety of existing standard libraries. This work demonstrates this functionality using MATLAB. We have developed the OpenFlexure MATLAB Client7 to enable researchers to perform automated imaging experiments seamlessly. Many students and researchers are familiar with MATLAB, and its image and video analysis tools make it powerful software for automated microscopy. This server-client architecture also means that one client can control several microscopes, by establishing connections to their independent IP addresses. Following the OpenFlexure ethos, the code is fully open source.
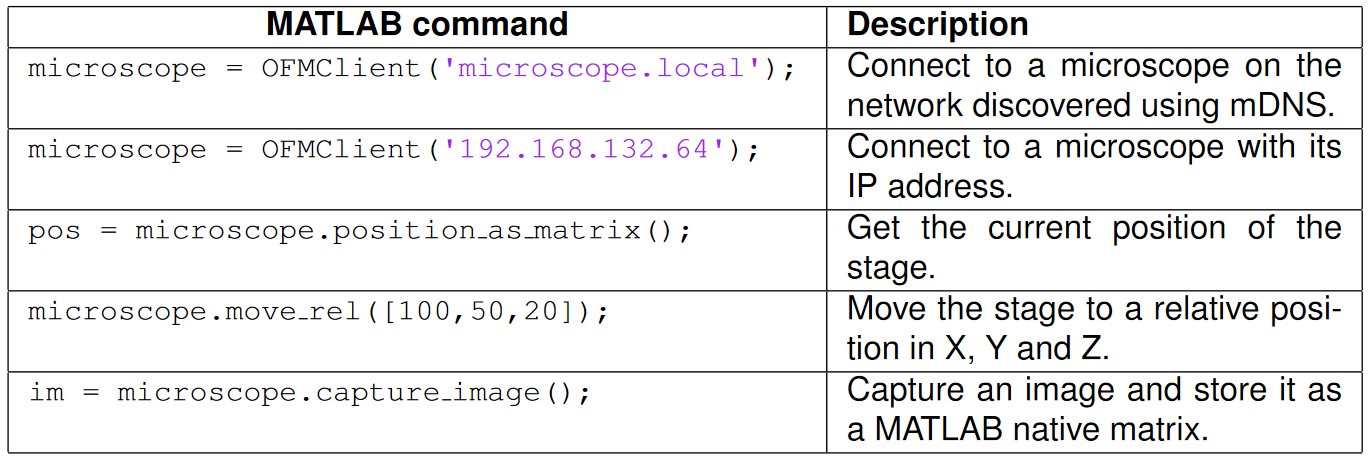
To use the client, users download the files from the MathWorks File Exchange8 and add them to the MATLAB path. The set of classes and functions in these files abstract the complexity of creating HTTP requests away from the end user, allowing them to use intuitive native MATLAB functions and data structures. A few examples of the MATLAB commands are given in Table 1.Table 1: Example commands for controlling the OpenFlexure Delta Stage from a MATLAB script.
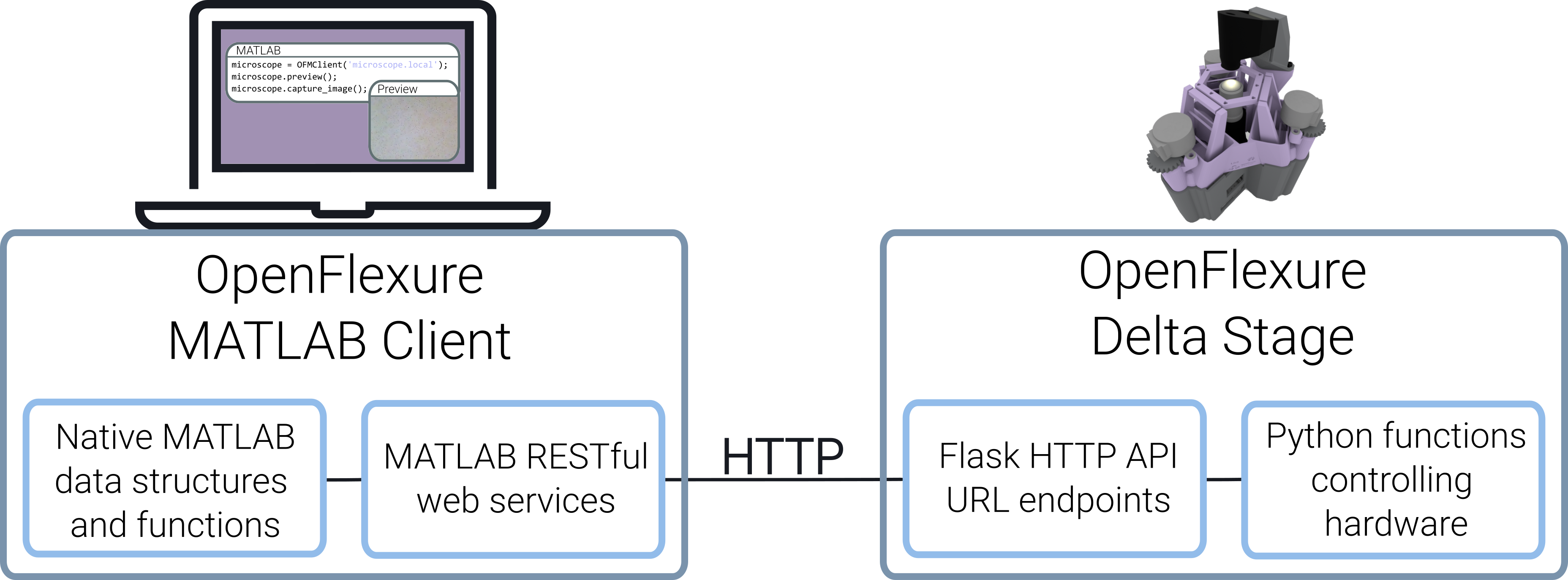
Figure 2: The MATLAB client runs on any computer with a MATLAB installation. It can control several OpenFlexure Delta Stages via HTTP commands.
As illustrated in Figure 2, the MATLAB OFMClient class contains core functions for operating the microscope. These functions convert the MATLAB native data structures into HTTP requests and send the payload to the API URLs. When a response has been received from the microscope server, they convert the response into the correct MATLAB data structure. The classes and functions are fully documented using the MATLAB help functionality.
If the microscope server has extensions installed, then their API URLs are separate to the core function URLs. However, when the MATLAB Client first connects to the Delta Stage, it requests a list of extensions installed on the microscope, and their associated API URLs. Users can then use their extensions directly from MATLAB in the same way as the core API.
To demonstrate the potential of the OpenFlexure MATLAB client, we use an example of Differential Dynamic Microscopy (DDM)9. This video analysis technique uses a video captured by a microscope to calculate the dynamic properties of colloids in soft materials, or biological objects such as bacteria and cells. It is an automated technique that does not require any user-setting of parameters. To conduct this experiment, the user prepares a sample of colloids of unknown size suspended in a known fluid. The sample is loaded into the Delta Stage. The user starts MATLAB on their computer and connects the microscope to the computer with Wi-Fi or an ethernet cable. The user establishes the connection to the microscope using the MATLAB client classes and a preview video from the camera can then be shown. The user's script can command the microscope's stage to move to a position and run the auto-focus routine. The command can be sent from the MATLAB script to record a video of a certain length with a desired framerate. Once the video has finished recording, it is automatically sent back to the user's computer. The MATLAB script can then analyse the video using the DDM technique and calculate the colloid's size based on their dynamic properties.
As both the video acquisition and analysis are now automated from a single MATLAB script, more complicated experiments can be performed. The video acquisition could occur over several time intervals, for example to measure the relaxation time of non-Newtonian fluids. The script could move the stage to different positions, and videos could be recorded at the boundaries and centre of the fluid. For high throughput, parallelized biological experiments, it is possible to control several OpenFlexure Delta Stages from a single MATLAB script.
The OpenFlexure MATLAB client transforms the OpenFlexure Delta Stage into an automatable device suitable for complex experiments. Its small size means that it can be kept on a laboratory bench or in an MSC, whilst its low manufacturing cost means that it opens automated microscopy to labs all over the world.- References
- openflexure-delta-stage [Repository], 2021. [Online]. Available: https://gitlab.com/openflexure/openflexure-delta-stage.
- J. T. Collins, J. Knapper, J. Stirling, J. Mduda, C. Mkindi, V. Mayagaya, G. A. Mwakajinga, P. T. Nyakyi, V. L. Sanga, D. Carbery, L. White, S. Dale, Z. Jieh Lim, J. J. Baumberg, P. Cicuta, S. McDermott, B. Vodenicharski, and R. Bowman, “Robotic microscopy for everyone: the OpenFlexure microscope,” Biomedical Optics Express, vol. 11, no. 5, p. 2447, 2020.
- J. T. Collins, J. Knapper, J. Stirling, S. McDermott, and R. Bowman, “Modern Microscopy with the Web of Things: The OpenFlexure Microscope Software Stack,” 2021. [Online]. Available: http://arxiv.org/abs/2101.00933.
- M. Kovatsch, R. Matsukura, M. Lagally, T. Kawaguchi, K. Toumura, and K. Kajimoto, Web of Things (WoT) Architecture, 2020. [Online]. Available: https://www.w3.org/TR/2020/REC-wot-architecture-20200409/.
- microscope-extensions [Repository], 2021. [Online]. Available: https://gitlab.com/openflexure/microscope-extensions.
- A. Neumann, N. Laranjeiro, and J. Bernardino, “An Analysis of Public REST Web Service APIs,” IEEE Transactions on Services Computing, vol. 1374, no. c, pp. 1–14, 2018.
- OpenFlexure Microscope MATLAB Client toolbox [Repository], 2021. [Online]. Available: https://gitlab.com/openflexure/openflexure-microscope-matlab-client.
- OpenFlexure Microscope MATLAB Client, 2021. [Online]. Available: https://uk.mathworks.com/matlabcentral/fileexchange/86478-openflexure-microscope-matlab-client.
- R. Cerbino and V. Trappe, “Differential dynamic microscopy: Probing wave vector dependent dynamics with a microscope,” Physical Review Letters, vol. 100, no. 18, pp. 1–4, 2008.