LifeHack: An Open-Source, Modular Microscope for Live & Fixed Cell Single Molecule Imaging
- Abstract number
- 203
- Presentation Form
- Poster
- Corresponding Email
- [email protected]
- Session
- Poster Session 3
- Authors
- Josh Edwards (1), Kevin Whitley (1), Sudheer Peneti (1), Yann Cesbron (1), Seamus Holden (1)
- Affiliations
-
1. Newcastle University
- Keywords
Open-source, super-resolution, single molecule, autofocus, microscopy, fluorescence, STORM
- Abstract text
Summary:
The LifeHack microscope is an open-source modular microscope capable of Live & Fixed Cell Single Molecule Imaging. The LifeHack offers performance beyond both current commercial and open source microscopes including live 3D drift correction and is easily modifiable to keep pace with the changing demands of an advanced microscopy lab. Comprehensive microscope designs have been released through a dedicated website (https://holdenlab.github.io/LifeHackWebsite/). This includes complete CAD models and detailed instructions for building and alignment of the system and use of the custom software.
Introduction:
Single molecule microscopy can reveal both sub diffraction limited structures and molecular dynamics directly in living cells but, unsurprisingly, requires advanced microscopes to do so. Commercial microscopes that provide these capabilities are expensive and inflexible. Home built systems are almost infinitely modifiable but require significant expertise and time to create. Open-source microscopes reduce this initial setup barrier while retaining the flexibility of home built systems. However, open-source systems currently lack advanced features for live cell microscopy such as dedicated incubation enclosures and focus lock.
To address this we have produced the LifeHack open source microscope, which is capable of high performance live and fixed single molecule imaging, and for use as a platform for further microscopy development.
Methods:
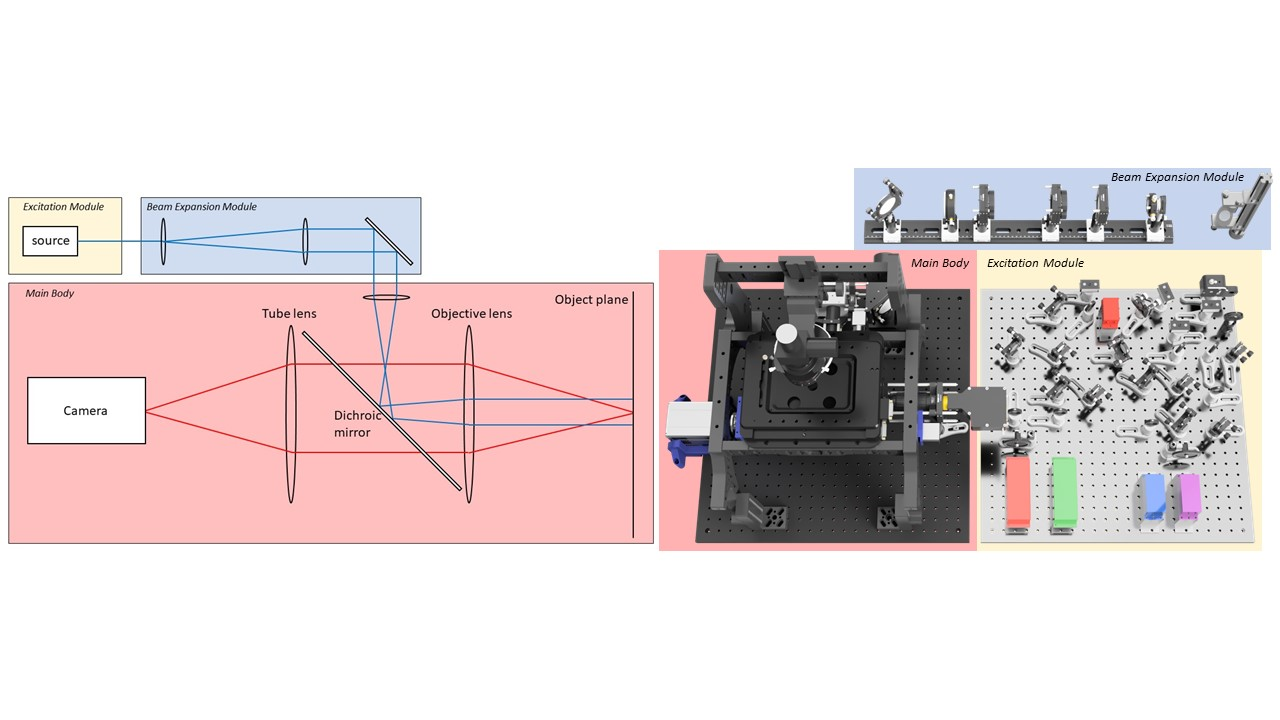
The entire microscope has been designed in 3D CAD software (Autodesk Fusion 360). Microscope CAD files are available on the LifeHack website (https://holdenlab.github.io/LifeHackWebsite/). The microscope contains 3 main modules and 3 sub-modules (Figure 1-3), which can be used together or separately adapted for integration into other systems. The complete parts list can be purchased for approximately £150,000.
Modules:
- Excitation module: Lasers, AOTF and electronic beam shutter.
-
Beam Expansion module: Raises and expands excitation beam. Open beam design for maximum power. - Main Body: Includes components for ring TIRF1, 3D single molecule localization2 and hardware autofocus systems
Figure 1: Left) Schematic of a basic fluorescence microscope. Right) Modular breakdown of the LifeHack microscope.
Sub-modules:
- Incubation Box: A custom acrylic box enclosing the main body. Includes large hatches for sample and hardware access.
- FocusShifter: Reflection based autofocus system with 10μm lock range.
- ImLock: Real time closed loop 3D drift correction by infrared-brightfield imaging.
Results/Discussion:
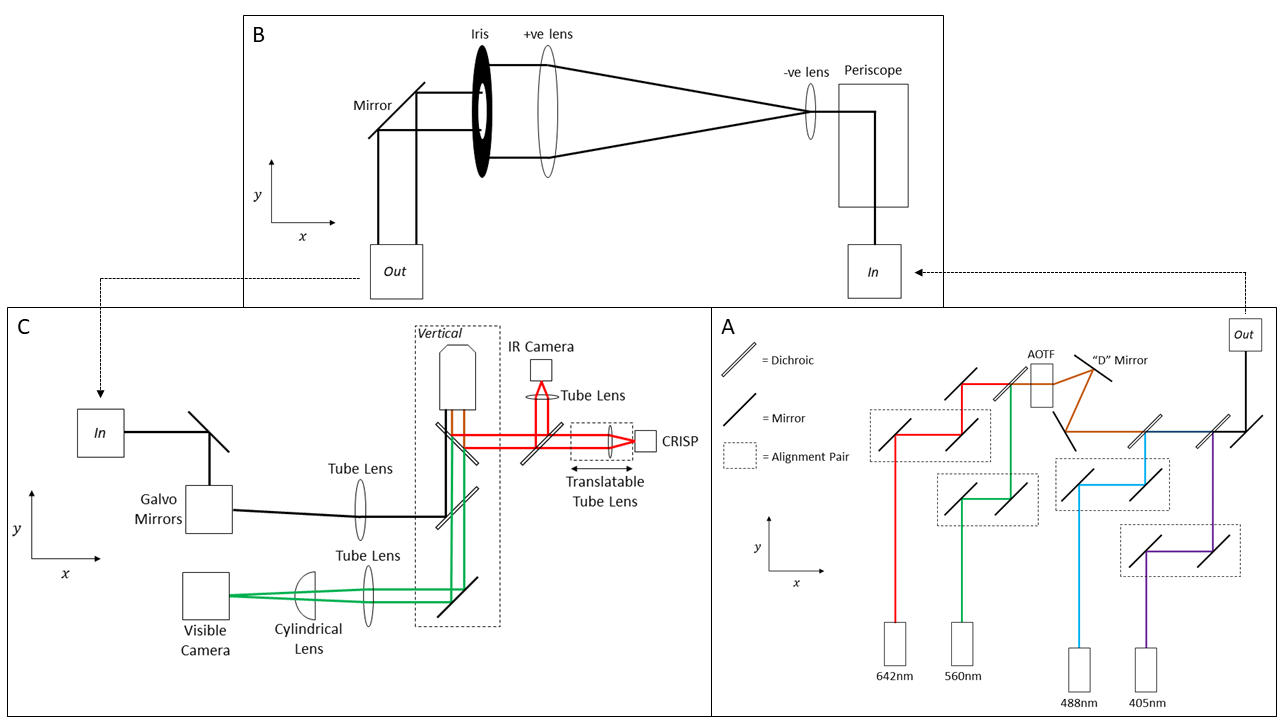
Figure 2: Optical diagram of the LifeHack microscope. A) Excitation module. B) Beam Expansion module. C) Main Body.
In the LifeHack we have produced a system that acts as a high performance live cell single molecule microscope out of the box and also acts as a microscopy development platform. The core features of the microscope are discussed below.
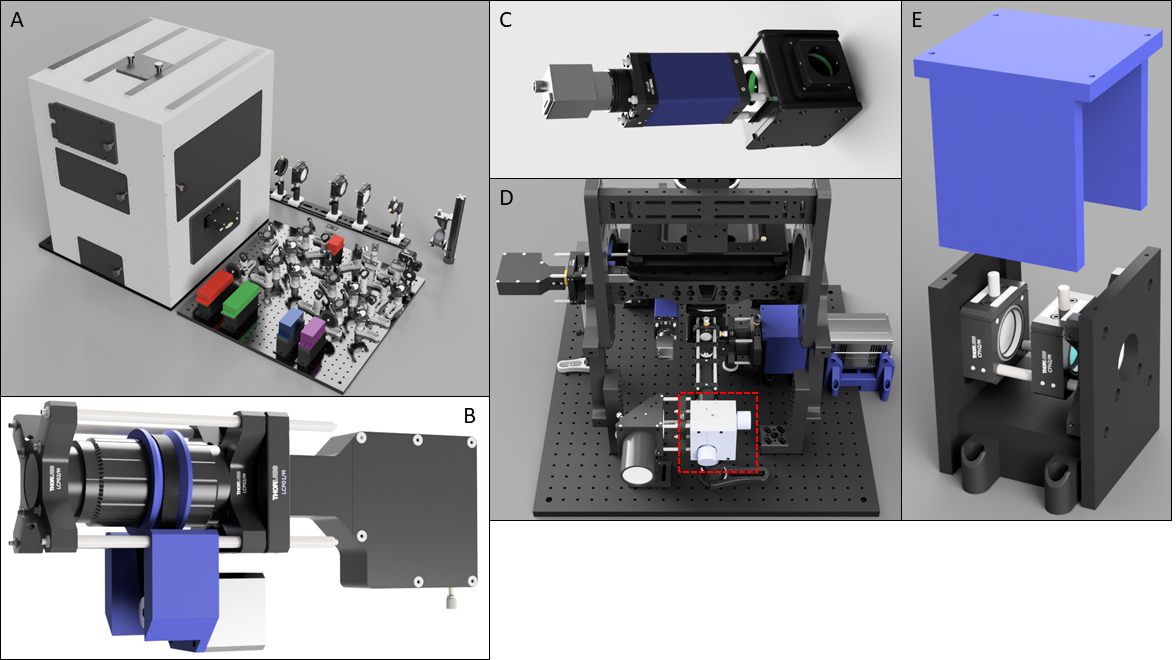
Microscope enclosure. We designed a custom heated microscope enclosure to maintain the entire microscope and sample at physiological temperatures (Figure 3A). This incorporates multiple access hatches for sample manipulation and microscope alignment.
Closed loop drift correction. Axial and lateral drift strongly degrade resolution of single molecule imaging. To address this, we designed two open hardware solutions for high precision closed loop drift correction. Reflection based autofocus systems maintain precise axial position within a 500 nm – 1000 nm distance from a glass-water interface. We designed the FocusShifter (Figure 3B), which extends the lock range of reflection based autofocus to approximately 10μm, allowing imaging of internal cellular structures.
We designed the ImLock (Figure 3C) to provided infra-red image based stabilisation across all three dimensions with a z range of approximately 50μm3. The system operates via cross-correlation of infrared brightfield images acquired simultaneously with fluorescence measurements, with high precision drift correction performed by closed loop feedback control of the microscope stage. The stability that this provides enables long imaging sequences such as for STORM4 or live cell microscopy time-lapses to be acquired. The ImLock compliments the FocusShifter but can also replace it in samples where no clear glass-water interface exists.
Ring TIRF illumination. Background rejection is crucial in single molecule imaging. To provide this we incorporated Ring TIRF1 and HILO illumination using a pair of Galvo-scanning mirrors (Figure 3D). Through the course of a single image exposure the excitation beam illuminates the coverslip from all angles thereby reducing shadowing or artefacts caused by inhomogeneities in the sample.[SH4]
Figure 3: A) Render of full LifeHack microscope. B) Render of FocusShifter showing motorised lens tube. C) ImLock module. D) Rear view of Main Body showing galvo-mirrors. E) 3D printed astigmatic lens box with translation rail.
3D localization microscopy. Astigmatic 3D localisation2 is provided by a cylindrical lens positioned in a 3D printed box (Figure 3E) immediately before the camera. This contains a rail mounting system allowing for axial adjustment of the lens position to tune the astigmatism strength.
High power multi-colour illumination. Four high power lasers (100mW 405nm, 150mW 488nm, 300mW 561nm, 1W 642nm) provide sample illumination, using an open beam path for efficiency. Therefore the system is capable of simultaneous 2 channel and sequential 4 channel microscopy with sufficient power for STORM4 imaging.
Open-source hardware. Open-source designs must be comprehensively documented to enable their adoption. To this end we have released a website (https://holdenlab.github.io/LifeHackWebsite/) with complete 3D models of both the microscope as a whole and individual modules. We also extensively documented the build process to provide clear guidance for construction, alignment, and operation of the system.
Conclusion:
We have designed, constructed, and released a fully featured open source single molecule microscope. Comprehensive designs and instructions for both building/alignment of the system and use of the custom software are provided. In comparison to existing open-source microscopes, the LifeHack is particularly advantageous for live cell imaging. This is due to its stable, super-resolution microscopy compatible body, precise temperature control and robust hardware autofocus/live drift correction in all 3 dimensions over a large depth range.
We are continuing to actively develop this microscope with particular focus on the live drift correction modules. We are also currently characterising the performance limits of the system.
Microscopy and biophysics labs frequently demand more of a microscope than commercial systems can offer. We hope that the LifeHack will act as both a high performance microscope for demanding single molecule and super-resolution experiments while also serving as an easy to extend development platform, significantly decreasing overall microscope development time.
- References
1. Ellefsen, K. L., Dynes, J. L. & Parker, I. Spinning-Spot Shadowless TIRF Microscopy. PLOS ONE 10, e0136055 (2015).
2. Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 319, 810–813.
3. Whitley, K. D. et al. FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. (Zenodo, 2021). doi:10.5281/ZENODO.4574236.
4. Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).