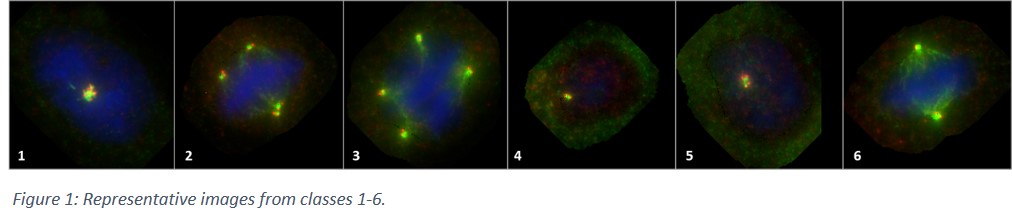
Development of a deep neural network based fully automated centrosome analysis workflow
- Abstract number
- 213
- Presentation Form
- Poster Flash Talk + Poster
- DOI
- 10.22443/rms.mmc2021.213
- Corresponding Email
- [email protected]
- Session
- Stream 2: Software and Smart Microscopy
- Authors
- Dr Gabor Pajor (1, 3), Dr Siegfried Hänselmann (2), Dr Barbara Waldkirch (2), Dr Rainer Will (1), Dr Andreas Plesch (2), Prof Alwin Krämer (1)
- Affiliations
-
1. Deutsches Krebsforschungszentrum
2. MetaSystems
3. Unversity of Pecs Medical School
- Keywords
centrosome, automated microscopy, deep neural network, DNN
- Abstract text
Chromosome instability - a major hallmark of cancer - is very often the consequence of centrosome amplification. Centrosomes are mostly visualized by immunofluorescence microscopy, but precise evaluation is error-prone and featured with inter-personal/-laboratory variance. A diagnostic and high-throughput screening methodology is thus highly desirable. Aim of our study was to develop a novel fully automated fluorescence light microscopy application, being able to determine the proportion of cells carrying aberrant centrosomes per sample. We are combining a deep neural network (DNN) classifier with an automated slide scanning system to generate a workflow that is able to process a large number of centrosome labelled slides with only minimal human interaction. Our project is a work in progress; current summary describes results of the almost finalized DNN to operate within the complete workflow.
MCF10A cells carrying different KRAS point-mutations (resulting in different level of centrosome amplification) were used for primary training of the DNN. Centrosomes were stained using immunofluorescence (antibodies were: CP110/568(red); γ-Tubulin/488(green)); nuclear counterstaining was performed using Hoechst 33342. From five different mutated MCF10A cell lines, 47 slides were stained, from which, more than a hundred and fifty thousand cells (156 699) were manually screened. Out of those, for training the DNN, more than thirty-five thousand (35 610) cells were categorised into seven (7) classes. These were (1) amplified centrosomes in interphase of the cell cycle; (2) amplified centrosomes in M-phase, featuring (pseudo)bipolar mitosis; (3) amplified centrosomes in M-phase, featuring multipolar mitosis; (4) non-amplified (i.e. normal) centrosomes in G1-phase; (5) non-amplified (i.e. normal) centrosomes in G2&S-phases; (6) non-amplified (i.e. normal) centrosomes in M-phase; (7) objects which cannot be classified into any of the above classes. Accordingly, it is also true, that classes 1-3 all belong to the bigger group of ‘amplified centrosomes’ whereas classes 4-6 represent cells with ‘non-amplified centrosomes’. Slides were scanned using Metafer Slide-Scanning System (MetaSystems Hard & Software GmbH, Altlussheim).
The resulting data set was divided into two non-overlapping subsets (training set and a validation set). The validation set consisted of a random subset of scans, including all images from these scans, such that each cell-class in the validation set contained around 10% of the total number of images of this class. Keras (version 2.2.4) [14] with TensorFlow (version 1.12.0) [15] as backend was used to train a custom convolutional neural net on the training set. In order to increase robustness against image variations and to avoid pure memorization of images, training images were continuously altered using various augmentation techniques such as image flipping, rotation, zooming and the addition of random noise. The training was performed on a single Nvidia Geforce GTX 1070 Ti graphics card.
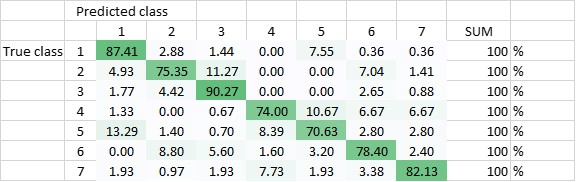
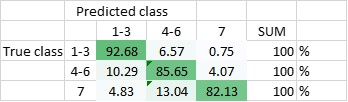
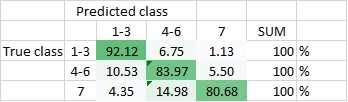
The DNN has been trained to be able to differentiate between the seven classes, and - in another setting - to be able to differentiate classes 1-3 (amplified centrosomes) from classes 4-6 (non-amplified centrosomes) and class 7. Currently, the DNN’s overall accuracy in predicting all of the classes correctly is 80,5% (Figure 2), whereas its ability to distinguish ‘amplified’ from ‘non-amplified’, is close to 90% (88,3%; Figure 3). The latter result was replicated when already the training of the DNN was done on a pooled data-set (Figure 4.; accuracy: 87,1%).
Further experiments are needed to fine-tune performance of the DNN before integrating it into the automated slide scanning system. This will result in a workflow which fully automatically analyses a sample, assessing the normal to aberrant ratio of centrosomes (including sub-classifying cells according to main cell cycle phases), with the only mandatory user interactions being the initial loading of the slide feeder and the final review of results.
Figure 2 Total accuracy of DNN - trained to recognize all seven classes (DNN1) - is currently 80,5%
Figure 3 Total accuracy of DNN1 to distinguish amplified centrosomes from non-amplified centrosomes is currently 88,3%
Figure 4 Total accuracy of DNN - trained with only three classes (DNN2) - is currently 87,1%
- References